Abstract
Waldenstrom's Macroglobulinemia (WM) is characterized by activating mutations in MYD88 and CXCR4 in in 93-95% and 30-40% of patient's, respectively. Functional analysis of these mutations has revealed downstream activation of NF-kB, through BTK/IRAK as well as AKT/ERK activation downstream of HCK and CXCR4. Transcriptome analysis using next generation sequencing demonstrated strong upregulation of BCL2 and overall dysregulation of the BCL2 family of genes.
Isoform based evaluation of splicing assumes that the data consists of a mixture of known isoforms, which may not be a reasonable assumption for a cancer transcriptome wherein mutational events and transcription factor dysregulation may cause unique combinations of splicing events not observed in healthy donors (HD). To explore alternative splicing in Waldenstrom's Macroglobulinemia (WM), we developed a MISO (Katz et al, Nat Meth 2010) based protocol to analyze next generation RNA sequencing data from 77 WM patients, 16 HD plasma cell samples, 9 HD CD19+CD27- peripheral blood B-cells and 9 HD CD19+CD27+ memory B-cells. Fifty-five patients harbored MYD88 mutations, 20 of whom also carried activating mutations in CXCR4. Twenty-two patients were wild-type (WT) for both genes, and was specifically enriched to insure adequate numbers for comparative study. To identify dysregulated splicing events, the memory B-cell cohort was merged and down-sampled for use as common comparator control for the WM and other HD samples. Estimating the variance in the control group is critical for statistical analysis so a jackknife permutation analysis where control samples pulled out and compared to the remaining samples was performed. This analysis was build into an automated pipeline based on two-pass STAR aligned bam files and corresponding tools to aggregate and analyze the data were developed using R. Novel intron retentions were investigated by identifying intronic regions not overlapping known exons. The number or reads mapping to these regions relative their flanking exons was calculated and compared.
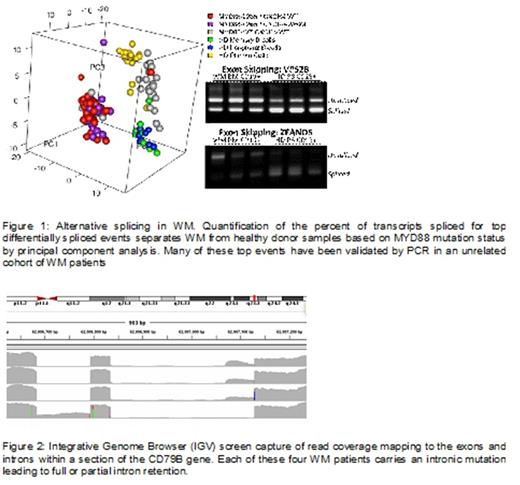
Filtering against events observed in the control permutation analysis and other healthy donor samples allowed for the identification of individual splicing events associated with WM. Splicing events were present in genes that regulated splicing itself (SF1, PTBP1), as well as pathways critical to WM growth and survival including NF-kB (ZFAND5, LYN, PKCD), ERK1/2 (CTBP1, VPS28, CCM2), and TP53 (PML, EIF4G2). PCR was successfully used to validate many of these splicing events (Figure 1). Preliminary intron retention analysis identified 4/77 (5%) of patients had intronic retentions on CD79B that were caused by intronic somatic mutations near the intron exon boundary (Figure 2). CD79B is a critical component of the B-cell receptor pathway and coding mutations have been observed in 7-15% of WM patients (Hunter et al, Blood 2013; Poulain et al, Am J Hematol, 2013).
The retentions identified in this study represent a novel form of CD79B mutation that has not been previously reported. Additional analysis and functional characterization of these events is planned. These studies aim to identify novel biomarkers and therapeutic targets for WM while developing analytical tools applicable to other malignancies.
Castillo: Janssen: Consultancy, Research Funding; Millennium: Research Funding; Pharmacyclics: Consultancy, Research Funding; Abbvie: Research Funding. Anderson: MedImmune: Membership on an entity's Board of Directors or advisory committees; Millenium Pharmaceuticals: Membership on an entity's Board of Directors or advisory committees; Oncopep: Other: scientific founder; Gilead Sciences: Membership on an entity's Board of Directors or advisory committees; C4 Therapeutics: Other: scientific founder; Bristol-Myers Squibb: Membership on an entity's Board of Directors or advisory committees. Treon: Pharmacyclics: Consultancy, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal